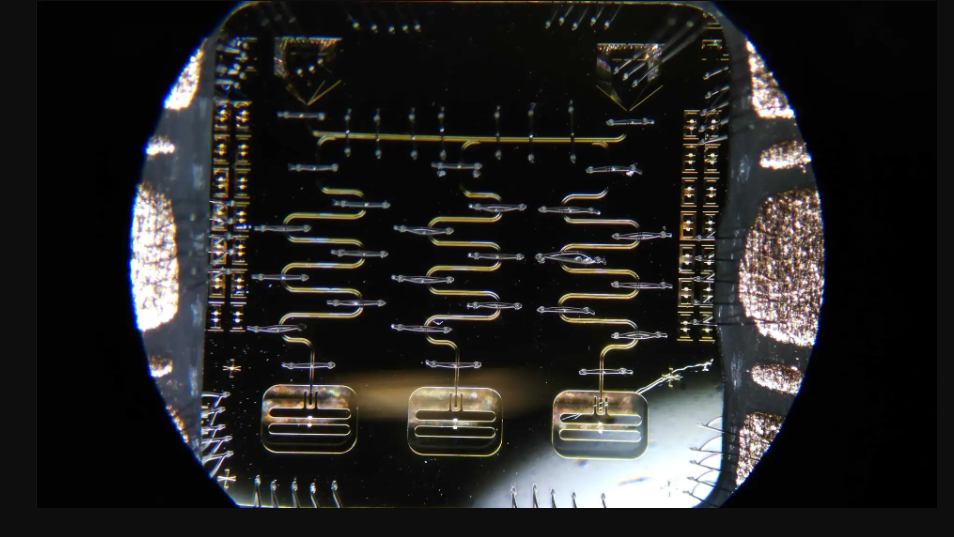
UMD Awarded $2 Million to Build a Quantum Biosensing Test Bed
- Details
- Category: Department News
- Published: Monday, November 18 2024 10:07
Physics Professor Wolfgang Losert, Cell Biology and Molecular Genetics Professor Kan Cao, Chemistry and Biochemistry Professor John Fourkas, and Electrical and Computer Engineering Associate Professor Cheng Gong were awarded $2 million by the U.S. Air Force Office of Scientific Research to build a test bed to study how neural networks process information and develop new approaches to quantum computing and sensing inspired by the living brain. As principal investigator of this multidisciplinary and cross-institutional project, Losert will collaborate with both UMD faculty members as well as other academic and industry partners to better understand and recreate the brain’s unique capacity for learning and adapting quickly—abilities that far surpass traditional computer systems. Wolfgang Losert
Wolfgang Losert
“The human brain is remarkable in how efficiently it can learn and process information. For example, we only need to touch a hot stove once to learn not to do it again,” Losert explained. “But current artificial intelligence systems need more than just that. Typically, they require enormous amounts of data and computing power to learn new tasks through numerous rounds of trial and error.”
While traditional computers process information through individual components working in sequence, the brain distributes information across many networks of cells working in parallel. This fundamentally different approach allows for faster learning and adaptivity but with far less energy consumption than a computer. Losert and his team hope to identify the biological mechanisms behind this efficient method of learning in the brain.
For this research, a key focus is on astrocytes, a type of brain cell that makes up more than half of the cells in the human brain. Long considered mere support cells for neurons, astrocytes are now recognized as crucial to how the brain processes information. By engineering laboratory-based systems that incorporate both neurons and astrocytes, Losert’s team will closely observe how the two types of cells form living neural networks and react when exposed to various types of stress like ultrasound or electrical fields.
Recent discoveries by the neuroscientist on Losert’s team, assistant research scientist Kate O’Neill, and other researchers have already shown that astrocytes actively participate in brain signaling and may be essential to the brain’s ability to both learn and adapt to new situations quickly. Further observations could provide insights into how the brain maintains its performance under different conditions and may lead to more resilient forms of artificial intelligence (AI).
“Interestingly, one aspect that makes biocomputing so unique—the multitude of different signals in living neural networks, such as electro-magnetic, chemical and mechanical signals—also opens up another exciting aspect of our work. We can use living neural networks to test and improve quantum sensors for a range of biomedical applications,” said Losert, who is also an MPower Professor and interim associate dean for research in the College of Computer, Mathematical, and Natural Sciences with a joint appointment in the Institute for Physical Science and Technology.
Quantum sensors have the potential to measure minute physical changes like the presence of magnetic fields or electrochemical activity in cells in minimally invasive ways. Novel non-invasive biosensors could allow scientists and health care professionals to observe brain processes in patients that they couldn’t see before, potentially leading to better medical treatments and a more nuanced understanding of brain performance.
With this award, Losert’s team aims to bridge the gap between artificial and biological computing systems and help create new technologies that combine the best features of both.
“By understanding and replicating how brain cells work together, we hope to create more efficient and adaptable computing systems,” Losert said. “This project represents the start of a new paradigm in biocomputing that may help shape the future of both AI and neuroscience.”
###
The grant will also facilitate collaborations with researchers from the U.S. Air Force Research Library, the National Quantum Laboratory (QLab), Lockheed Martin, the National Research Council of Italy (CNR) and the University of Bari Aldo Moro.